Quickstart Guide
Requirements
Conmo was developed under Python version 3.7.11 so it should work with similar or more recent versions. However, we cannot claim this to be true, so we recommend using the same version. To be able to use Conmo you need to have installed a Python interpreter and the following libraries on your computer:
If you want to make a contribution by modifying code and documentation you need to include these libraries as well:
We suggest to create a new virtual enviroment using the Conda package manager and install there all dependences.
Installation
The fastest way to get started with Conmo is to install it via the pip command.
pip install conmo
And then you will be able to open a Python interpreter and try running.
import conmo
Some TensorFlow warnings might come up if your computer doesn’t have installed a GPU, although that’s not a problem for running Conmo.
You can also install Conmo manually downloading the source code from the Github repository.
git clone https://github.com/MyM-Uniovi/conmo.git
cd conmo
Then if you haven’t prepared manually a conda enviroment, you can execute the shell-script install_conmo_conda.sh to install all the dependences and create a Conda enviroment with Python 3.7.
cd scripts
./install_conmo_conda.sh conda_env_name
If your operating system is MacOS, please, check Known issues & Limitations section for more information about compatibility of Conmo with Apple M1 and M2 CPUs.
If your operating system is not Unix-like and you are using Windows 10/11 OS you can create the Conda enviroment manually or use the Windows Subsytem for Linux (WSL) tool. For more information about its installation, please refer to Microsoft’s official documentation..
To check if the Conda enviroment is activated you should see a (conda_env_name) in your command line. If it is not activated, then you can activated it using:
conda activate conda_env_name
Overview
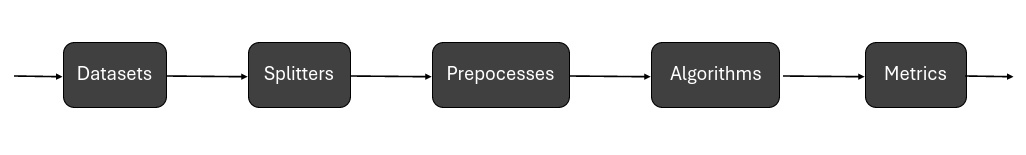
The experiments in Conmo have a pipeline-based architecture. A pipeline consists of a chain of processes connected in such a way that the output of each element of the chain is the input of the next, thus creating a data flow. Each of these processes represents one of the typical generic steps in Machine Learning experiments:
- Datasets
Defines the dataset used in the experiment which will be the starting data of the chain. Here the dataset will be loaded and parsed to a standard format.
- Splitters
Typically in Machine Learning problems the data has to be splitted into train data and test data. Also here you can apply Cross-Validation techniques.
- Preprocesses
Defines the sequence of preprocesses to be applied over the dataset to manipulate the data before any algorithm is executed.
- Algorithms
Defines the different algorithms which will be executed over the same input data stream (as a result of the previous stage). It can be one or several.
- Metrics
Defines the different metrics that can be used to evaluate the results obtained from the algorithms.
Further details and documentation about modules, functions and parameters are provided in the API Reference.
Running an experiment
Here is a brief example on how to use the different comno modules to reproduce an experiment. In this case with the predefined splitter of the Server Machine Dataset, Sklearn’s MinMaxScaler as preprocessing, PCAMahalanobis as algorithm and accuracy as metric.
Import the module if it hasn’t been imported yet and other dependences:
1from sklearn.preprocessing import MinMaxScaler 2 3from conmo import Experiment, Pipeline 4from conmo.algorithms import PCAMahalanobis 5from conmo.datasets import ServerMachineDataset 6from conmo.metrics import Accuracy 7from conmo.preprocesses import SklearnPreprocess 8from conmo.splitters import SklearnSplitter 9from sklearn.model_selection import PredefinedSplit 10from sklearn.preprocessing import MinMaxScaler
Configure the different stages of the pipeline:
1dataset = ServerMachineDataset('1-01') 2splitter = SklearnSplitter(splitter=PredefinedSplit(dataset.sklearn_predefined_split())) 3preprocesses = [ 4 SklearnPreprocess(to_data=True, to_labels=False, 5 test_set=True, preprocess=MinMaxScaler()), 6] 7algorithms = [ 8 PCAMahalanobis() 9] 10metrics = [ 11 Accuracy() 12] 13pipeline = Pipeline(dataset, splitter, preprocesses, algorithms, metrics)
Create an experiment with the configured pipeline. The first parameter is a list of the pipelines that will be included in the experiment It can be one or more. The second parameter is for statistical testing between results, but this part is still under development and therefore it cannot be used:
1experiment = Experiment([pipeline], [])
Start running the experiment by calling
launch()method:1experiment.launch()
As a result of the execution of the experiment a specific folder structure will be created in
~/conmo:
/dataThis directory contains the various datasets that have already been imported (downloaded and parsed) and are therefore already available for use. They are stored in parquet format for better compression. For each of the subdatasets included in each dataset, there will be a data file and a labels file.
/experimentsThis directory contains all the executions of an experiment in Conmo in chronological order. Each directory corresponds to an experiment and has in its name a timestamp with the time and day when this experiment was run. Within each experiment directory there will be another one for each pipeline, and within this one there will be as many directories as the number of steps each pipeline has been determined to contain These folders contain the input and output data used by each step of the pipeline. They are also stored in parquet format, in the same way as the datasets in the
/datafolder.